Every Job Needs the Right Tool
Oligonucleotide Therapeutics (ONTs) and mRNA-based vaccines have been actively researched for decades, and broad approval of RNA therapeutics, including ONTs, is in process. The early development problems of delivery, stability, and cellular uptake have an expanding list of solutions, such as the use of liquid nanoparticles (LNPs), GalNAc receptor targeting, and an ever-growing library of chemical modifications. However, the effects of intracellular distribution on ASO activity is not fully understood.2 In this context, the importance of new tools to quantify and localize ASOs and siRNAs cannot be overstated.
While valuable, the current methods for acquiring analytical data have their limitations. There is an urgent and pressing need to expand the toolbox for data collection to better ensure the safety and efficacy of drugs. This need is particularly acute given that the gold standard for nucleic acid quantification, qPCR, lacks sensitivity to heavily modified ONTs and their shorter metabolites.
Choosing the Right Wrench
Presently, there is a critical need to develop robust immunoassays to complement traditional methods, which is crucial for effectively monitoring the absorption, distribution, metabolism, excretion (ADME), immunogenicity, and in-situ localization and quantification of the growing variety of ONTs. This can be a difficult and time-consuming process since ONTs are not highly immunogenic and tend to have highly randomized nucleic acid sequences. LC-MS/MS, HI-ELISA, ISH, and qRT-PCR are standard assays to detect and quantify ONTs; at present, most studies on intracellular presence and trafficking rely on covalent attachment of a fluorophore onto the oligonucleotide 3 These methods are laborious and expensive when faced with multiple diverse nucleic acid sequences, chemical structures, and modifications, including conventional oligonucleotides (12-15 bases), DNA-RNA hybrids, PS-modified backbones, single-stranded oligonucleotide mixtures (cocktails), single-stranded RNA (ssRNA), double-stranded RNA (dsRNA), silencing RNA (siRNA), nucleotides, and nucleosides.
The development of new methods is not just a choice, but a necessity. These new methods are crucial to support traditional assays and fill the gaps that constitute their deficiencies. Orthogonal assays to quantify and visualize the uptake of ONTs and their transport throughout target cells and tissues should be developed with new, robust tools with the potential to revolutionize the industry. The utilization of immunoassays that deploy traditional antibody tools for generating data sets on par with qPCR and LC-MS/MS should be considered, as they have the potential to significantly advance our understanding of ONTs.
Next Generation Toolbox
By comparison, antibodies have shown the ability to detect post-translation modifications on proteins to elucidate the location and degree of modification. We took a similar approach to generate antibodies to common oligonucleotide modifications, such as PS, OME, and OMe, to detect ONTs independent of sequence or structure. Dual-labeling or multiplexing assays can visualize the localization of ONTs and their effect on biological processes. Monitoring transport organelles or downstream protein changes can demonstrate that novel ONTs are successfully entering target cells and tissues, as well as their desired therapeutic outcomes, and provide for quantification in biofluids.
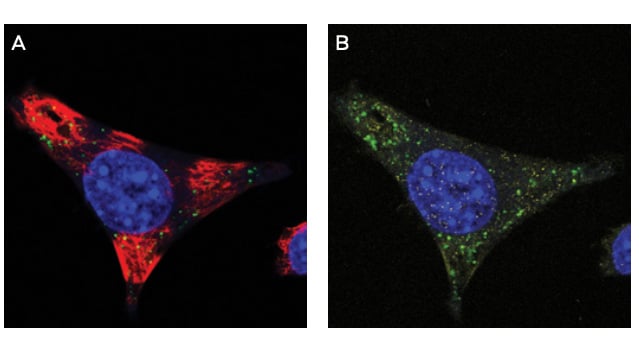
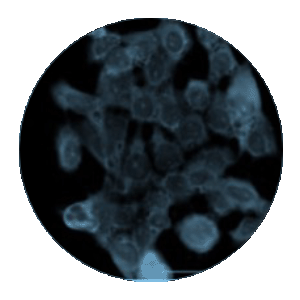
Figure 1. IF of Anti-PS monoclonal antibody clone PS05. Mouse glioma cells derived from C57 black mice were cultured and treated with ASO drug. After fixation with paraformaldehyde, cells were stained with a(red) and anti-PS antibody clone PS05 (green). Clone PS05 was used at a 1:2000 dilution. Punctate cytoplasmic staining is consistent with endosomal storage of ASO within the cell, as expected for this OTX drug. Vehicle only treated cells showed no staining (not shown).
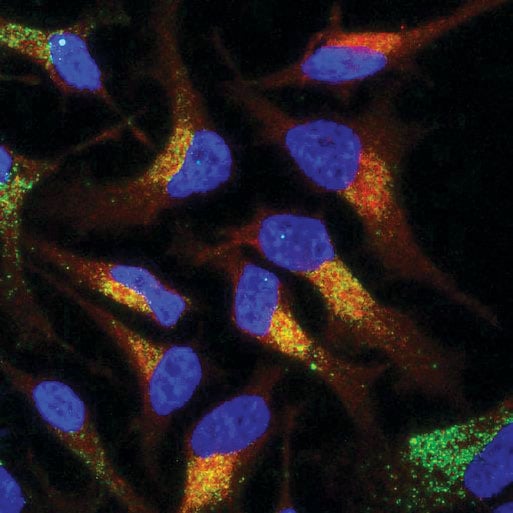
Figure 1. Co-localization of an antisense oligonucleotide (ASO) using Immunoflourescence Microscopy. A method for the localization of PS-modified oligonucleotides in HeLa cells has been developed for use. A 100% PS-modified MALAT1 specific 16-mer was detected using the ModDetect™ PS Panel. A 100 nM dose of ASO was delivered gymnotically and visualized using ModDetect. Dual imaging of the RAB9A endosomal marker indicates cellular uptake. (Image courtesy of Nucleic Acid Therapeutic Accelerator)
Quantifying and analyzing ONTs may also involve utilizing image analysis software to measure the intensity and distribution of ONT signals. This quantitative approach provides valuable insights into the relative abundance and spatial distribution of ONTs. To deepen the understanding of ONTs’ interactions, contextual analysis is employed to correlate ONT localization with histological or cellular features, revealing how ONTs engage with specific cellular processes or structures.
Understanding the spatial localization of ONTs is crucial for therapeutic development, as it helps optimize their efficacy and minimize off-target effects. It ensures that ONTs reach their intended targets effectively. Additionally, in pathological studies, analyzing ONT localization in disease models can provide valuable information on how ONTs influence cellular and tissue processes, enhancing understanding of disease mechanisms and treatment responses. Integrating ONT detection with antibody-based techniques thus enables researchers to obtain comprehensive insights into ONT localization and context, significantly advancing the development and application of ONT-based therapies.
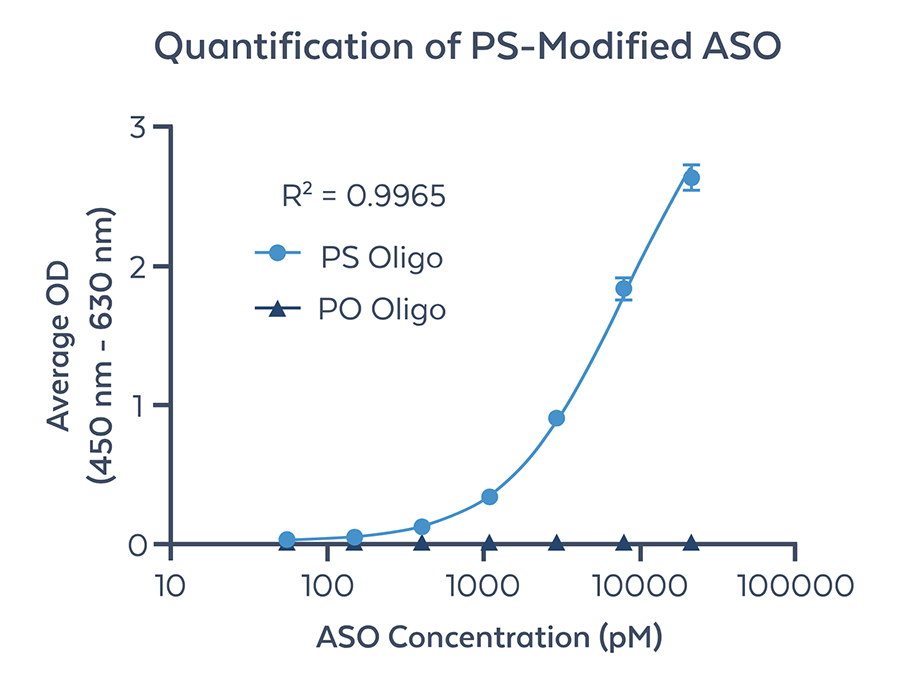
Figure 2. Quantification of fully PS modified ASO by ELISA using anti-PS monoclonal antibody (clone PS04). Non-modified PO oligonucleotide of the same sequence was used as a negative control (PO Oligo). The curve was generated using PS-modified ASO diluted in buffer from 55 pM to 21 nM in concentrations and subsequently detected using anti-PS monoclonal antibody. A standard curve was plotted as the average OD result versus the log of ASO concentration in pM using a 4PL best fit formula. The lower limit of quantitation (LLOQ) as defined as the lowest standard detected is 55 pM. The limit of detection (LOD) was determined to be <11pM based on standard definitions of the term. The upper limit of quantitation (ULOQ) as shown in 21 nM. Further optimization and sensitivity enhancement will likely increase the LLOQ by one log unit.
Rockland’s ModDetect™ Antibody Panels can be used to provide a sensitive and quantifiable detection method for ONTs in various matrices, tissue lysates, and biofluids. Subsequent visualization techniques, such as immunofluorescence (IF) and immunohistochemistry (IHC), can be developed in parallel using the versatility of ModDetect panels to assess the biodistribution and localization of the target ONTs. In addition, the use of ELISA for ModDetect will provide supportive, robust data to support future ONT development.
One Tool-Fits-All Solution
Immunoassays have a well-documented record of generating comprehensive data sets to characterize new therapeutics. There is a growing need for robust antibody tools to facilitate immunoassay development that supports the discovery of new oligonucleotide therapies and pre-clinical ONT candidates. Rockland’s ModDetect Panels have shown the ability to detect ONTs of different sizes and quantities of modification. Additionally, these same reagents can detect the ONT targets in multiple assay types. The versatility and performance of these reagents represent a new tool to support the demands of a maturing therapeutic field.